This content originally appeared on HackerNoon and was authored by Image Recognition
Table of Links
Materials and Methods
2.1 Vector Database and Indexing
Discussion
4.1 Dataset and 4.2 Re-ranking
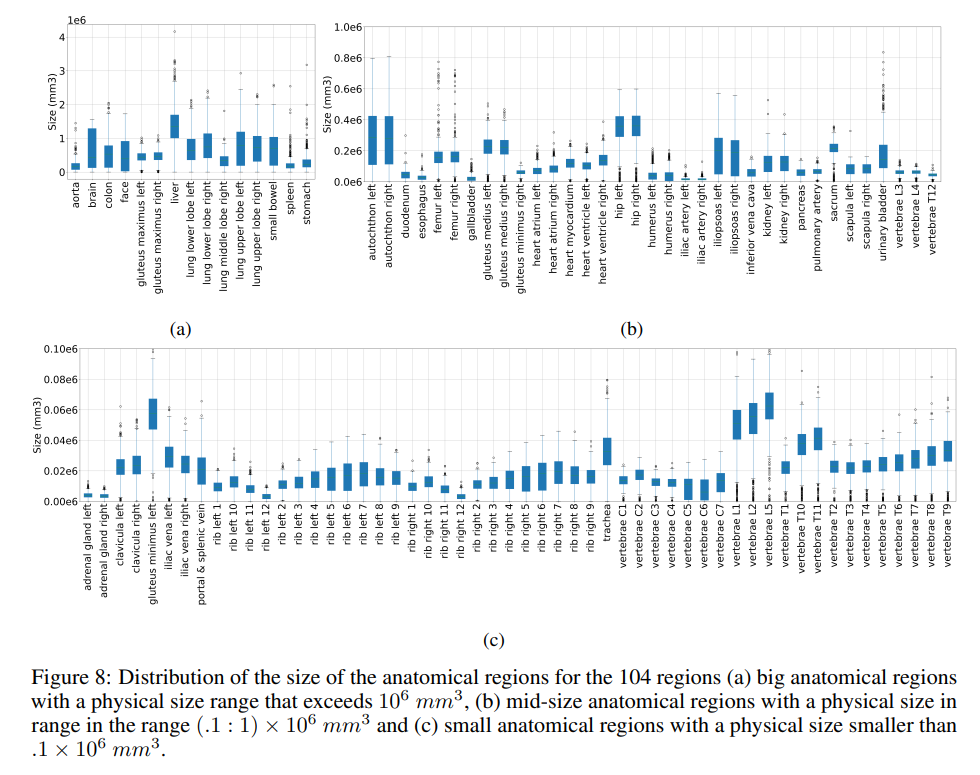
4.4 Volume-based, Region-based and Localized Retrieval and 4.5 Localization-ratio
4.3 Embeddings
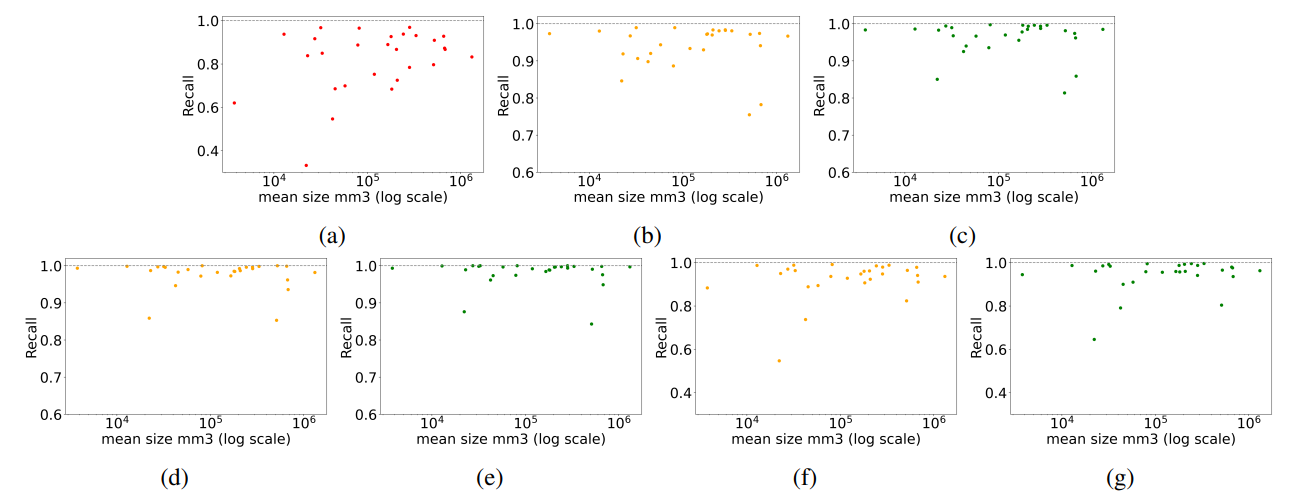
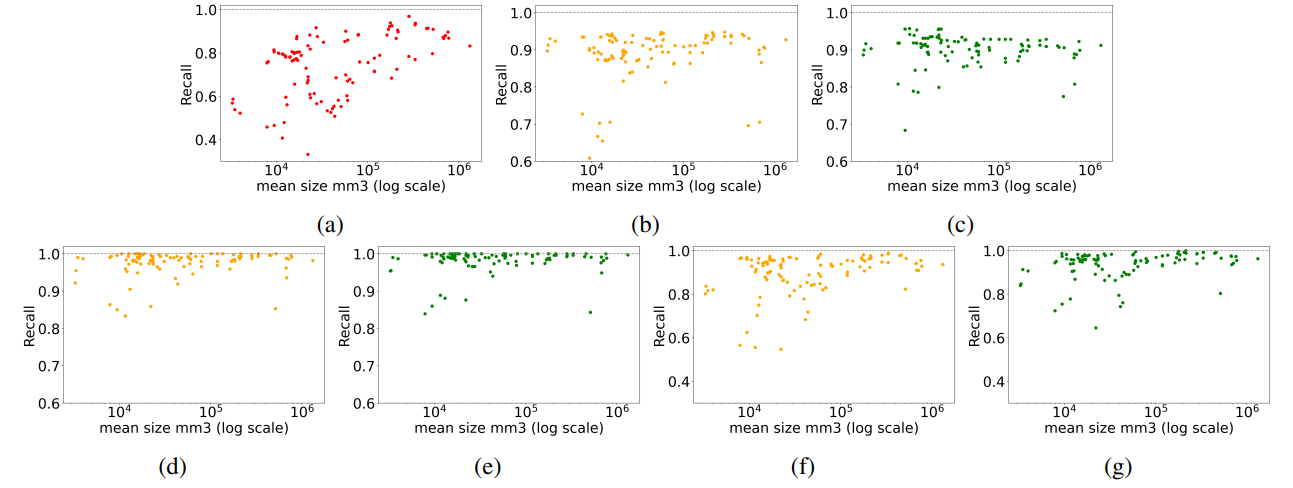
It was shown that embeddings generated from self-supervised models are slightly better for image retrieval tasks than those derived from regular supervised models. This is true for coarse anatomical regions with 29 labels (see Table 20) as well as fine-granular anatomical regions with 104 regions (see Table 21). This is roughly preserved for all modes of retrieval (i.e. slice-wise, volume-based, region-based, and localized retrieval). More generally, the differences in recall across differently pre-trained models (except pre-trained from fractal image) are very small. Practically, the exact choice of the feature extractor should not be noticeable to a potential user in a downstream application. Further, it can be
\
\ concluded that pre-training on general natural images (i.e. ImageNet) resulted in slightly more performant embedding vectors than domain-specific images (i.e. RadImageNet). This is unexpected and subject to further research.
\ Although, the model pre-trained of formula-derived synthetic images of fractals (i.e. Fractaldb) showed the lowest recall accuracy the absolute values are surprisingly high considering that the model learned visual primitives out of rendered fractals. This is very encouraging as the Formular-Driven Supervised Learning (FDSL) can easily be extended to very high number of data points per class and also several virtual classes within one family of formulas [Kataoka et al., 2022]. Additionally, the mathematical space of formulas for producing visual primitives is virtually infinite and thus it is the subject of further research whether radiology-specific visual primitives can be created that outperform natural image-based pre-training. Again, FDSL does not require the effort of data collection, curation, and annotation. It can scale to a large number of samples and classes which potentially results in a very smooth and evenly covered latent space.
\ Embeddings derived from DreamSim architecture showed the highest overall retrieval recall in region-based and localized evaluations. DreamSim is an ensemble architecture that uses multiple ViT embeddings with additional finetuning using synthetic images. It is plausible that an ensemble approach outperforms single-architecture embeddings (i.e. DINOv1, DINOv2, SwinTransformer, and ResNet50). Therefore, the usage of DreamSim is currently the preferred method of embedding generation.
\ Worth discussing is an observation that can be found in all tables presenting recall values. Across all model architectures (column) there are usually a few anatomies or regions (i.e. row) that show lower recall on average (see "Average" column). For example, in Table 2 "gallbladder" showed poor retrieval accuracy, whereas in Table Table 4 "brain" and "face" showed lower recall. The observation of isolated low-recall patterns can be seen across all modes of retrieval and aggregation. The authors of this paper cannot provide an explanation, as to why certain anatomies perform worse in certain retrieval configurations but gain high recall in many other retrieval configurations. This will be subject to future research.
\
\
\
:::info Authors:
(1) Farnaz Khun Jush, Bayer AG, Berlin, Germany (farnaz.khunjush@bayer.com);
(2) Steffen Vogler, Bayer AG, Berlin, Germany (steffen.vogler@bayer.com);
(3) Tuan Truong, Bayer AG, Berlin, Germany (tuan.truong@bayer.com);
(4) Matthias Lenga, Bayer AG, Berlin, Germany (matthias.lenga@bayer.com).
:::
:::info This paper is available on arxiv under CC BY 4.0 DEED license.
:::
\
This content originally appeared on HackerNoon and was authored by Image Recognition
Image Recognition | Sciencx (2025-08-29T07:00:03+00:00) DreamSim and the Future of Embedding Models in Radiology AI. Retrieved from https://www.scien.cx/2025/08/29/dreamsim-and-the-future-of-embedding-models-in-radiology-ai/
Please log in to upload a file.
There are no updates yet.
Click the Upload button above to add an update.
